From DNA to Genetic
Genealogy
Everything you wanted to
know but were afraid to ask
Stephen P. Morse
This article first appeared in the Association of
Professional Genealogists Quarterly (March 2009).
It was reprinted in Selected Lectures on Genealogy: An Inroduction to Scientific Tools (2013).
For the record, let me state that I am not a geneticist, not a biologist, and not a chemist. And I have no affiliation with a DNA testing laboratory. I’m an engineer. So why am I writing a paper on genetic genealogy and what are my qualifications for doing so?
I’m writing this because DNA has become a hot topic in genealogy lately. Unfortunately most of the DNA lectures being presented at genealogy conferences assume that the listener knows the basics. I didn’t have that knowledge because DNA was “invented” after I went to school. I knew a little about genes, having studied Mendel’s laws in high-school biology. And I remembered that chromosomes determined sex. But beyond that I was lost -- I didn’t even know how genes related to chromosomes.
So I decided to teach myself about genes, chromosomes, and DNA. While doing so, I realized that I was not alone – most genealogists were as confused as I was. So I developed a lecture (and subsequently this paper) based on what I learned and how I learned it. It is my hope to give you the background you’ll need to be able to understand the other DNA lectures.
1. INTRODUCTION (GENES, CHROMOSOMES, AND DNA)
The field of genetics originated in 1865 with Gregor Mendel and his pea experiments. One hundred and sixty-five years later it evolved to the point where it could be useful to genealogists, as exemplified by Megan Smolenyak’s coining of the term genetealogy in the year 2000.
Everything we need to know about genetealogy involves three basic concepts – genes, chromosomes, and DNA. The relation between them is simple:
Traits are determined by GENES.
Genes are located on CHROMOSOMES.
Chromosomes are composed of DNA.
Now let’s look at each of them in more detail.
a. Genes
Mendel is credited as the father of genetics. But he never actually saw a gene, nor did he even use that term. Instead he postulated the existence of discrete entities and drew conclusions about them based on statistical observation from experiments involving the breeding of pea plants. In particular, he concluded that:
Each trait is determined by a pair of discrete entities (genes).
One gene of each pair comes from each parent.
The two genes are not blended; instead one of them dominates.
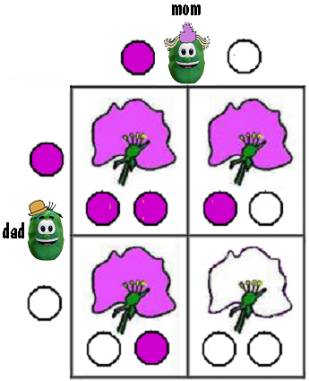
As an example, let’s consider the color of the pea plant’s flower. The flower can be either purple or white. The color is determined by a pair of genes, each of which specifies either purple or white, where purple is dominant.
If the child pea inherits a purple gene from each parent pea, the child’s flower would be purple.
If the child inherited a white gene from each parent, the child’s flower would be white.
If the child inherited a purple gene from one parent and a white gene from the other, the child’s flower would be purple because purple is dominant. And it would not matter which parent contributed the purple gene and which the white gene.
This is illustrated in the figure below. The circles represent the genes and each box represents a possible child.
b. Chromosomes
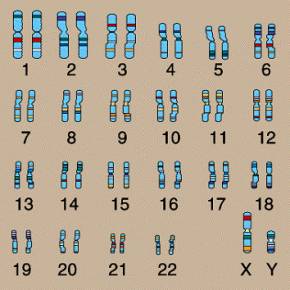
In the nucleus of nearly every human cell is a set of 46 chromosomes. These chromosomes are numbered in pairs from 1 to 22 for a total of 44. Each of the two remaining chromosomes are either X or Y. The numbered chromosomes are referred to as autosomes, and the X and Y chromosomes are called sex chromosomes. The chromosomes are shown in the following figure.
The X and Y chromosomes are called the sex chromosomes for an obvious reason – they determine the sex. Specifically:
Males have one Y chromosome and one X chromosome.
Females have two X chromosomes.
One sex chromosome comes from each parent.
What about two Y chromosomes? Since women are XX, perhaps real men are YY. Let’s consider that.
Assume that the original man, Adam, was YY. The original woman, Eve, was of course XX. Their children, who would receive one sex chromosome from each parent, would all be YX and therefore all males. And, indeed, the children of Adam and Eve were Cain, Abel, and Seth – all males. There is no way that Adam and Eve could have produced any daughters (XX).
We all know what happened next. Cain killed Abel, leaving Abel with no descendants. Cain became an outcast, and we certainly don’t want to be descended from him. So the future of the whole human race depended on Seth. But how?
Seth turned to Eve for help. This was the original Oedipus complex. It was also the world’s first cover-up – there is no mention of it in any history book, biology book, or theology book. Seth and Eve together produced XX daughters and YX sons. There is no way for YY to ever be produced again. So if YY ever did exist, it died out, never to be seen again.
c. DNA
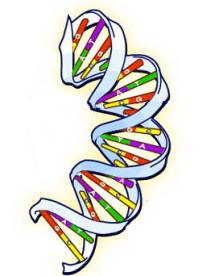
A chromosome is a long Deoxyribo Nucleic Acid molecule, better known as DNA. The DNA structure is a double helix with the two strands running in opposite directions (see figure below). The direction of each strand is determined by its chemical composition, but we won’t say any more about the direction since it doesn’t affect the genealogy story.
Each of the strands consists of four repeating compounds, which are called bases or nucleotides. The four bases are:
Adenine (A)
Cytosine (C)
Guanine (G)
Thymine (T)
Fortunately we don’t have to remember these names but can refer to them simply by their initial – A, C, G, T.
Sometime you might see Uracil (U) instead of Thymine (T). This occurs in RNA. I mention this so that you won’t be confused if you should see mention of base pairs A, C, G, and U. But we don’t have to know anything about RNA as far as understanding genetic genealogy. So forget I ever said Uracil.
The bases on the two strands always pair up in a specific way. In particular, A on one strand always pairs with T on the other, and C always pairs with G. Why are there no other pairings, such as A with G? That’s due to the geometry of the compounds – their shape is such that the only pairings possible are A-T and G-C. The other pairings simply do not fit together.
d. Historical Perspective
To get a better understanding of the relation between genes, chromosomes, and DNA, let’s look at what happened when.
1865: Mendel discovers genes, but he is not believed. He presents his findings at a scientific conference, but is laughed at. After all, what does a monk know about biology?
1882: Under a microscope, Flemming sees little squiggly things in the center of a cell. He calls them chromosomes.
1900: deVries notices similarities between Mendel’s particles and the newly-discovered chromosomes. Finally Mendel’s name has been cleared but it is too late – he’s already dead.
1924: Microscopic studies show that DNA is present in chromosomes.
1952: Biologists Hershey and Chase realize that it is DNA that encodes the genes. The gene is determined by the values of the DNA base pairs in the gene.
1953: Watson and Crick describe the DNA double-helix structure, although arguably the credit should have gone to Franklin and Wilkins. This was partially rectified when the Nobel prize was awarded to Watson, Crick, and Wilkins (Franklin died four years earlier).
e. Relating Genes, Chromosomes, and DNA – a summary
Chromosomes are composed of DNA.
A gene is a subset of the DNA sequence having an identifiable function.
The DNA in between the genes is called junk DNA.
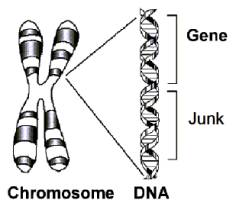
Going down the DNA sequence of a chromosome we encounter gene, junk DNA, gene, junk DNA, etc. In between each gene is junk.
Junk DNA – that’s an interesting concept! What is it? It is a DNA sequence for which no function has yet been identified. Perhaps it contains driftwoods from our evolutionary past, such as for fur color or tail length.
f. By the Numbers
The actual number of genes or base pairs is not important for genealogy. But as an engineer, I like to know if we are talking about two or two trillion. So here are the numbers, just to put things in perspective.
Base Pairs
How many base pairs are there in a gene?
Average length of a gene is 27 thousand base pairs.
Largest known gene has 2.4 million base pairs.
How many base pairs are there in a chromosome?
Depends on the chromosome number (see chart below).
Between 50 million and 250 million.
Chromosomes are numbered by decreasing number of base pairs (size places).
How many base pairs in all 46 chromosomes?
About 3 billion.
So we can completely identify a person by giving a sequence of 3 billion base pairs. No two people (except for clones or identical twins) would ever have the same 3 billion base-pair sequence.
Genes
How many genes in a chromosome?
Depends on the chromosome number (see chart below).
Between 200 and 3,000
How many genes in all 46 chromosomes?
When I first looked this up, it was thought to be 30,000.
Now that number is believed to be closer to 20,000.
I have no idea what it will be by the time you are reading this paper.
|
Chromosome |
Bases |
Genes |
|
1 |
245,203,898 |
2,968 |
|
2 |
243,315,028 |
2,288 |
|
3 |
199,411,731 |
2,032 |
|
4 |
191,610,523 |
1,297 |
|
5 |
180,967,295 |
1,643 |
|
6 |
170,740,541 |
1,963 |
|
7 |
158,431,299 |
1,443 |
|
8 |
145,908,738 |
1,127 |
|
9 |
134,505,819 |
1,299 |
|
10 |
135,480,874 |
1,440 |
|
11 |
134,978,784 |
2,093 |
|
12 |
133,464,434 |
1,652 |
|
13 |
114,151,656 |
748 |
|
14 |
105,311,216 |
1,098 |
|
15 |
100,114,055 |
1,122 |
|
16 |
89,995,999 |
1,098 |
|
17 |
81,691,216 |
1,576 |
|
18 |
77,753,510 |
766 |
|
19 |
63,790,860 |
1,454 |
|
20 |
63,644,868 |
927 |
|
21 |
46,976,537 |
303 |
|
22 |
49,476,972 |
288 |
|
X |
152,634,166 |
1,184 |
|
Y |
50,961,097 |
231 |
g. Example of Gene: Human Eye Color
Let’s consider eye color since we all probably recall from high-school biology that genes determine eye color. Specifically we were taught that brown eyes are dominant over blue eyes, so a person would have to have two blue-eyed genes in order to have blue eyes. What we weren’t taught (because it wasn’t known at that time) is where these genes are and what base values they have.
We now know the rest of the story. In the case of eye color, the controlling gene is the OCA2 gene, located on the long half of chromosome 15 in bands 1,1. As far as our genealogy goes, it’s not important that we know the name of the gene or where it is. But it’s fascinating to realize that geneticists have mapped out all the chromosomes and know not only where a particular gene is, but also the values of the base pairs in the gene.
In our OCA2 gene, there are 344,433 base pairs but only three of them are relevant for eye color. The location of those three are shown in the chart below, along with the values they would have for a blue-eyed person and the values for a brown-eyed person.
|
rs7495174 |
T (blue) |
C (brown) |
|
rs6497268 |
G (blue) |
T (brown) |
|
rs11855019 |
T (blue) |
C (brown) |
2. CRACKING THE DNA CODE (MAKING PROTEINS)
The DNA code is not important as far as our genealogy is concerned. It is important for understanding genetic diseases, which some genealogists are concerned with. If you are interested only in knowing who might be your second cousin, you can skip this section.
However this is the part of DNA that I find the most fascinating. I can’t help but see the similarity between the DNA code and a computer program. To realize that each cell in our body contains a computer program for building our body is truly amazing!
Recall that I’m not a biologist, so some of these points I am accepting simply on faith. For example, I really don’t know much about proteins. But the biologists tell us that every function in a living cell depends on proteins. So I will take that as gospel, and realize that if I knew the rules for making proteins, I would understand all the functions in our bodies.
Geneticists discovered that each gene is an instruction manual for making one protein. And the instruction manual is written in the language of DNA. To understand how to read the manual, we have to know what proteins are composed of. The biologists tell us that each protein is a sequence of amino acids. OK, I can accept that, even though I’m not sure what an amino acid is. The biologists then tell us that there are only 20 amino acids, as shown in the chart below.
|
Alanine |
Ala |
A |
|
Leucine |
Leu |
L |
|
Arginine |
Arg |
R |
|
Lysine |
Lys |
K |
|
Asparagine |
Asn |
N |
|
Methionine |
Met |
M |
|
Aspartic
acid |
Asp |
D |
|
Phenylalanine |
Phe |
F |
|
Cysteine |
Cys |
C |
|
Proline |
Pro |
P |
|
Glutamic
acid |
Glu |
E |
|
Serine |
Ser |
S |
|
Glutamine |
Gln |
Q |
|
Threonine |
Thr |
T |
|
Glycine |
Gly |
G |
|
Tryptophan |
Trp |
W |
|
Histidine |
His |
H |
|
Tyrosine |
Tyr |
Y |
|
Isoleucine |
Ile |
I |
|
Valine |
Val |
V |
The amino acids have names such as Asparagine, Isoleucine, and my all time favorite – Phenylalanine. These are names that only a biologist could love. If they were named by a computer scientist, they would probably be called Amino Acid 1.0, Amino Acid 1.1, etc. The biologists realized that we were not going to like those names, so they gave us a three-letter abbreviation for each amino acid. And in case we still found it cryptic, they gave us a one-letter abbreviation for each as well. Of course it’s kind of confusing to see that the one-letter abbreviation for Lysine is K, but if we stick with the abbreviations and forget the name, we’ll be OK.
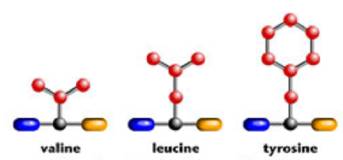
Since a protein is a sequence of amino acids, if we chain several amino acids together in a particular order we will make a particular protein. For example, here are the chemical structures for three of the amino acids:
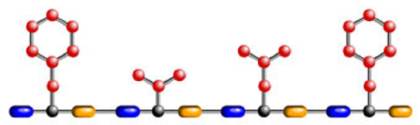
And here is a protein made from a sequence of those amino acids.
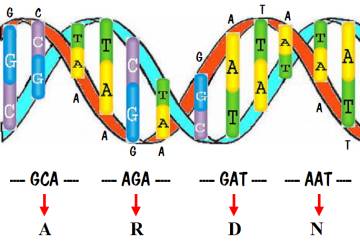
Now we are ready to talk about the DNA code. Recall that the DNA in each of our cells consists of a sequence of about 3 billion base pairs. What was discovered is that within a gene, every three pairs (called a codon) specifies one amino acid. This is shown in the following diagram. Note that the DNA sequence starts off with the codon GCA, and that specifies amino acid A. The next codon is AGA, specifying amino acid R. Then comes GAT specifying D, and AAT specifying N. An entire gene specifies a sequence of amino acids, which is a protein.
So we need to know the amino acid associated with every possible sequence of three base pairs. That information is shown in the chart below.
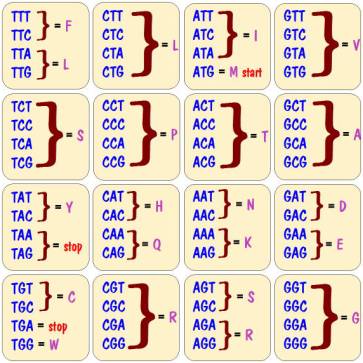
Note that there can be more than one codon specifying the same amino acid. For example, codon AGT and codon AGC both specify the amino acid S.
Note also that there are four special entries in the chart. Three of them, TAA, TAG, and TGA do not specify any amino acids but rather serve as a stop code. They are saying that you have reached the end of a gene, and there are no more amino acids in the protein. The fourth, ATG, denotes the amino acid M, and also serves as a start code saying that you are at the beginning of a gene. This of course implies that all proteins start with the amino acid M.
Below is an example of a DNA sequence for making a protein. We read this by going through the sequence looking for a start codon – ATG. When we find that, we know we are at the start of the gene and the first amino acid is M. Everything preceding the ATG is junk DNA. Next we see the codon CTG, and that specifies the amino acid L. We continue scanning the bases, three at a time, until we get to a stop codon. In this case, the stop codon that we get to is TGA. That tells us that we have reached the end of the gene, and there are no more amino acids for this protein. The bases after that are junk DNA.

3. THE BIRDS AND THE BEES (HOW WE INHERIT CHROMOSOMES)
Usually when we talk of inheritance, we are talking about what is passed from parent to child when the parent dies. In genetics, inheritance refers to what is passed from parent to child when the child is born. We will look at two kinds of inheritance – autosome inheritance and sex-chromosome inheritance. And we will learn how both of these can be used to tell us something about our ancestors, and therefore our genealogy.
a. Autosome Inheritance
We’ll first look at autosome inheritance. Recall that the autosomes are the numbered chromosome pairs – 1 to 22. Let’s consider chromosome number 7 as an example. Since a child inherits one number 7 chromosome from each parent, and since each parent has two number 7 chromosomes, it would be natural to think that the father passed one of his number 7 chromosomes directly to the child and the mother did the same. This is depicted in the following figure.
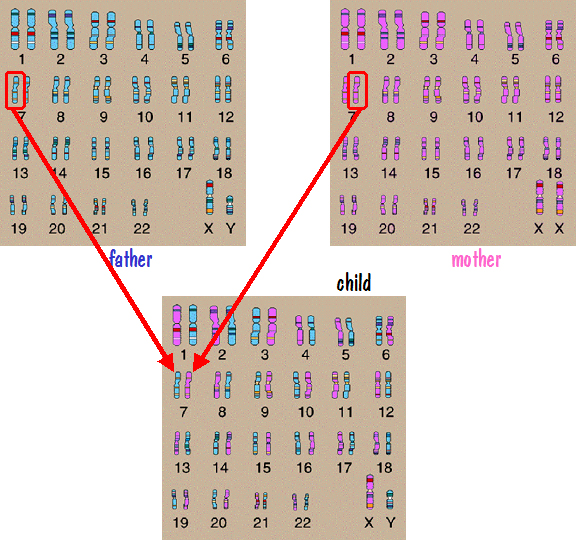
If this were the case, then the number 7 chromosome that the father passed would have come from one of his parents, say his mother. And the mother would have gotten the number 7 chromosome that she passed from one of her parents, say her father. So the child would have genetic information on number 7 from his paternal grandmother and his maternal grandfather. But he would have nothing from his other two grandparents. The genes on those grandparents number 7 chromosomes would be lost to all future generations descending from this grandchild.
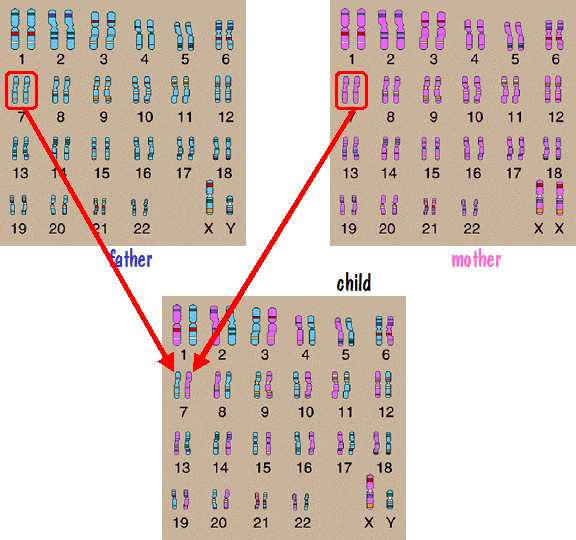
However that is not the case. Instead of the father passing one of his number 7 chromosomes intact, his two number 7 chromosomes are shuffled together forming a new chromosome that gets passed down. The mother does the same. So the child receives one number 7 chromosome from each parent, but each of those chromosomes is a mixture of the parent’s two number 7 chromosomes. By this means, genetic information on all the autosomes from all the grandparents get passed down. This is shown in the following figure.
b. Sex Chromosome Inheritance
Now for sex chromosome inheritance. Every cell in the mother’s body has two X chromosomes. However when the mother’s body produces an egg, the egg has a single X chromosome. Similarly, every cell in the father’s body contains an X and a Y, but the sperm cells his body produces contains a single sex chromosome – either an X or a Y. The child then receives an X from the mother and either an X or a Y from the father. This is depicted in the following two diagrams.
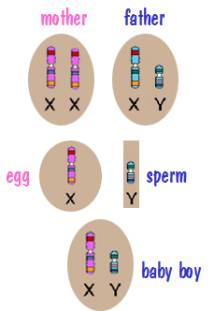
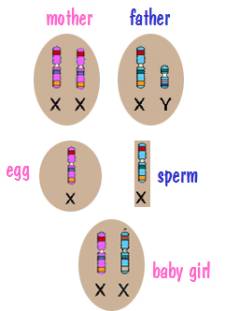
In both cases, the X chromosome that the child receives from the mother is a shuffled X chromosome. But the Y chromosome that the son receives from the father is an intact Y chromosome since the father had only one. Similarly the X chromosome that the daughter receives from the father is an intact X chromosome.
4. WHO’S YOUR DADDY (MUTATIONS ARE GOOD)
We can now utilize our knowledge of chromosome inheritance to determine if two people share a recent common ancestor, and are therefore not-too-distant cousins. We can also use it to determine something about our early ancestors who migrated from someplace in Africa, and can determine if our ancestor turned left or right when he got to Kenya. Such distant ancestry won’t help us construct our family tree, but some people like to know these things.
a. Like Father, Like Son
The key to doing this sort of genetic genealogy lies in the way the sex chromosome is inherited. The X chromosome involves some shuffling within the past one or two generations. But the Y chromosome is never shuffled – it is always passed from the father to the son intact. So the DNA code on the son’s Y chromosome should be identical to the father’s, and every male should have the identical Y chromosome to Adam. In that case we could do DNA testing and determine that we are all descendants of Adam. Although this might help us weed out the extra-terrestrial aliens among us, it would be of little help in constructing our family trees.
But fortunately mistakes happen, and the DNA is not always passed intact. Such mistakes are called mutations. Considering the vast number of cells in our body (in excess of ten trillion), mutations are probably happening all the time. But if any one cell mutates, it probably never gets noticed. The exceptions is in the sex cells (egg and sperm), because such a mutation would get passed down from parent to child.
So thanks to mutations, the Y chromosome passed from father to son will sometimes change, and the son’s Y chromosome might differ slightly from the father’s Y chromosome. It is this difference that will allow us to find out if two people are related, and how close the relation is.
There are two kinds of mutations that we are most interested in. They are referred to as SNiPs and STiRs. We will discuss each of these separately.
b. SNiP
SNiP is an acronym for Single Nucleotide Polymorphism. Those are big words, but the important word is Single. It refers to a mutation in which a single base (recall that another name for base was nucleotide) changed its value. So a DNA sequence of TGAT at a particular location in the DNA sequence (called a marker) might become TGCT when passed to the next generation.
A SNiP is an extremely rare event. It is so rare, that if it occurs at a particular marker to any person at any time in history, the probability of it ever occurring again at the same marker to any other person would be zero. That is, if the mutation occurred to one man who lived 75,000 years ago, no other man in all of history would ever mutate at that same marker. And, for the same reason, once the mutation occurs and gets passed down to future generations, it will never get undone.
So how do SNiPs tell us about our ancestors? Well if SNiPs are so rare, and if you and I both have the same SNiP value at a particular marker, then we must both be descended from that man in which the SNiP occurred 75,000 years ago. This is not going to tell us if we are second cousins or not. But it will tell us if we descended from the tribe in Africa that turned left when it got to Kenya thousands of years ago.
Where in the DNA sequence shall we test for SNiPs? What would happen if it occurs inside the DNA sequence of a gene? It could be a so-called silent mutation. That is, the resulting amino acid would be unchanged (recall that there could be more than one DNA sequence, all making the same amino acid), and that would have no effect on the organism. But, more likely, it would result in a different amino acid and therefore a different protein. This could have a profound effect on the organism, and the organism might not even be viable. So it would make no sense to look for mutations in genes in order to find our ancestors -- people having such mutations probably won’t live long enough to have descendants. On the other hand, if the SNiP occurs in the junk DNA, it will have no affect what-so-ever. For this reason, genealogists are interested in testing for SNiPs in junk DNA only.
c. STiR
STiR is an acronym for Short Tandem Repeat. Again we can ignore all these words except one – Repeat. It refers to a DNA sequence that repeats itself several times. Due to a mutation, the number of times it repeats could change, either upward or downward. For example, the DNA sequence GA[CTA][CTA][CTA][CTA][CTA]GT might mutate to GA[CTA][CTA][CTA][CTA][CTA][CTA]GT. The brackets have been added for emphasis, and they show that the sequence CTA has gone from five repetitions to six repetitions. The repeated sequence usually involve between 2 and 10 bases.
Unlike SNiPs, STiR mutations are not at all uncommon. They happen on average once in every 500 events. That means if two people have the same STiR value at a particular marker, they probably have a common ancestor within the last 500 generations. Well that means that they are cousins, but quite distant. If they have the same STiR values on more than one marker, we can reduce that number considerably and might be able to deduce that their common ancestor was only a few generations back. So the STiR values can be used to find out if two people are related within the timeframes of interest to genealogists.
For the same reason as SNiPs, the STiRs tested to determine relationships are in the junk DNA regions and not in the genes themselves. In fact, a STiR mutation in a gene is even more likely to result in a non-viable organism because it could change not just one amino acid (which is all a SNiP could do) but all the following amino acids in the sequence.
d. Nomenclature
The standard nomenclature for Single Nucleotide Polymorphism found in the literature is SNP. And it is usually followed with a parenthetical comment saying “pronounced snip.” Well if it is pronounced that way, I decided to spell it that way as well, using a small “i” to indicate that it is not part of the acronym. You won’t see it written as SNiP anywhere else but here – elsewhere you will see it written as SNP.
The standard nomenclature for Short Tandem Repeat found in the literature is STR. And that’s it – there is no comment about how to pronounce it because it is never pronounced as a word. Instead it is always spoken of as the three letters – S-T-R. That made no sense to me, and I decided to refer to it as STiR in this paper and pronounce it that way in my lectures. But you won’t see STiR written or pronounced that way anywhere else.
e. Who’s your Mommy?
Up to now, we have examined the direct male line based on the Y chromosome. Something similar can be done with the direct female line.
The 46 chromosomes are in the nucleus of each cell. Surrounding the nucleus are the energy bars of the cell – the mitochondria. Like the chromosomes, the mitochondria are composed of DNA, called mitochondrial DNA or simply mtDNA.
The sex cells (egg and sperm) are no exception, and they too contain mitochondria. So it would seem that a child should get mitochondria from both parents. However the mitochondria is in the tail of the sperm cell. The sperm that actually penetrates the egg works so hard to do so, and it barely manages to get its head in – the tail breaks off and never makes it. So the child gets its mitochondria from its mother only. (In a few extremely rare cases, paternal mitochondria has actually been found in an offspring).
The mitochondria is passed from a mother to all her children. But only her daughters pass that mitochondria to their children. Her sons children will get their mitochondria from their mother. That means that every woman received her mitochondria from her mother who in turn received it from her mother, and so forth.
If not for mutations, all women would have mtDNA identical to Eve’s. But mistakes do happen, and mtDNA mutates. The mutations are of the SNiP type, and occur very infrequently. So they can be used for tracing early migrations (did your mother turn left or right when she got to Kenya) and are not that useful for determining if two women have a common great grandmother.
f. Heinz 57 Varieties
We’ve seen above how Y-chromosome DNA can be used to learn something about your father’s father’s father’s father’s father, and mtDNA can tell you something about your mother’s mother’s mother’s mother’s mother. But you have 30 other great great great grandparents and you certainly don’t want to ignore them.
Recall that the autosomes (the numbered chromosomes) are not inherited intact but rather are a shuffling of the chromosome pair from each parent. So DNA from all your ancestors going all the way back is present in the autosomes. That means that testing of the autosomal DNA can give you some statistics on your mix of ancestors.
For example, the results of an autosomal test might say that you are 50% European, 25% East Asian, 12.5% Sub-Saharan African, and 12.5% Native American. That would mean that one of your parents was 100% European, and the other was a mixture of 50% East Asian, 25% Sub-Saharan African, and 25% Native American. That mixed parent would have one parent who was 100% East Asian and the other who was a mixture of 50% Sub-Saharan African and 50% Native American. And the latter would have one parent who was 100% Sub-Saharan African and 100% Native American (that must be the Indian Princess that we all hope to find in our family trees). This is illustrated in the following diagram.
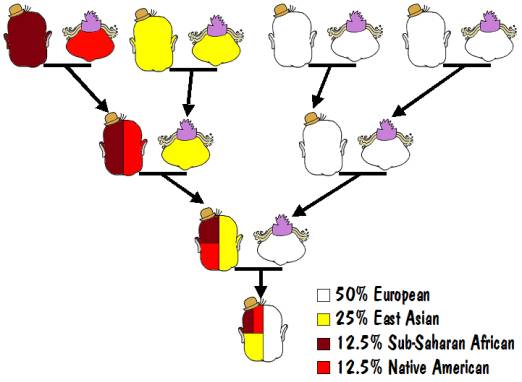
It should be mentioned that the percentages obtained from such autosomal testing are not very reliable, and many of the major testing laboratories don’t even offer this service.
5. OUT OF AFRICA (THE ROAD WE TRAVELED)
In the beginning God created man. And all men lived in Africa and had
identical Y chromosomes.
And in the year 60,000 BP (before the present), a
mutation occurred. One man was born
with a different base value at marker m91.
And he was shunned by the others and forced to leave the tribe. So he started his own tribe and called it
Haplogroup A. His tribe remained in
Africa, primarily around Ethiopia.
And then another man in the original tribe was born with
a mutation, and he too was asked to leave.
He started the BR tribe.
Then in the year 50,000 BP, two mutants appeared in the
BR tribe. The first started the B tribe
and became the pygmies. The second
started the CR tribe and immediately had two mutant sons, and they started
tribes C and DE. The C tribe left
Africa and went to Australia and the surrounding areas (Japan, Siberia, North
America).
And so this continued, and the tribes D through R appeared. And each tribe went off in a different direction to populate the earth.
|
---------------
A BR
--------------------
B CR
--------------------------------------
C DE F
--------------------------------------
G H IJ K
-------- -------------------------------
I J L M NO P
--------- --------
N O Q R
The preceding is a dramatization of course. But the facts about the mutations (all of them being SNiPs) and the haplogroups are true. And below is a chart (small and hard to read) showing the migration paths taken by the various groups.
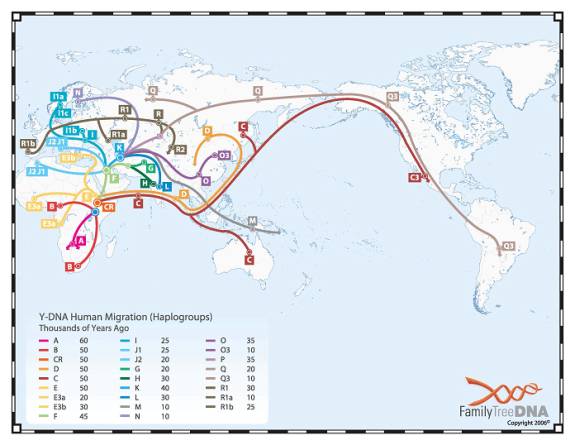
How do we know all this? By obtaining DNA from various indigenous groups around the world, and looking at their SNiP values.
Note that the mutations that define the tribes (haplogroups) are SNiPs. The standard DNA testing that many of us do involve STiRs because we are more interested in finding our second cousins than in knowing what route we took through Africa. Yet when we get back the results of our DNA testing, it usually tells what haplogroup we are in. How can they determine that from STiRs? They do so by having a large database of people for whom they’ve tested both SNiPs and STiRs. From the SNiPs, they know for certain which people are in which haplogroups. Then they examine the STiR values of the people in each haplogroup, and from that they are able to determine the probability that a particular set of STiRs will be in a particular haplogroup. Of course you can find out for certain what haplogroup you are in by having your SNiPs tested as well as your STiRs. But that costs more money.
The above migration pattern is based on the Y-chromosome mutation and tells you about your paternal lineage. Similar results can be obtained for the migration of your maternal lineage based on mtDNA mutations. An interesting conclusion from the maternal migration is that all European women living today are descended from only seven matriarchs who lived between 10,000 and 45,000 years ago. This finding has been popularized in a book titled “Seven Daughters of Eve.” Another finding is that all Ashkenazi Jewish women living today are descended from only four matriarchs.
6. EXAMPLES (GENGHIS KHAN, THOMAS JEFFERSON, ANASTASIA)
The following examples illustrate how Y-chromosome and mtDNA analysis have been used in forensic applications.
a. Genghis Khan and the Mongolian Empire
Eight Hundred years ago Genghis Khan conquered a region from the Pacific Ocean to the Caspian Sea. His armies massacred the males and raped the females in the conquered lands. Khan himself always got first pick of the females. Khan’s eldest son had 40 sons. His grandson had 22 legitimate sons and he added 20 virgins a year to his harem. Harems and concubines were the norm, and the males were very prolific. All told, this made for an excellent Petri dish in which to spread Khan’s Y chromosome.
Contemporary DNA testing indicates that 1 in 12 men (or about 16 million men) in the region of the former Mongolian Empire carry a common Y chromosome – a subtype of haplogroup C3. Furthermore, it has been established that this lineage of Y chromosome started about 1000 years ago. And it is found in only one population outside of the Mongolian Empire, namely Pakistan, where they have an oral tradition of being descended from Khan.
The conclusion is that we have isolated Khan’s Y chromosome, although the original mutation predates him by several generations. So it probably started with his great great grandfather or thereabout. Of course we’ll never know for sure if this is Khan’s Y chromosome unless we can find his body and test it. But the evidence is overwhelming.
b. The Thomas Jefferson Affair
In 1802 James Callender, disgruntled because Jefferson wouldn’t appoint him as Postmaster General, alleged that Jefferson fathered several children with Jefferson’s slave Sally Hemings. Jefferson’s policy was to offer no public response. However Jefferson’s daughter Martha denied the report, and two of Martha’s children maintained that Jefferson’s nephews, Peter and Samuel Carr, were the father.
Let’s look at the Jefferson family tree and see if Y-chromosome testing will help here. Jefferson himself had only daughters (except for a son who died before he was even named), so we won’t be looking for any of Jefferson’s direct male descendants. Instead we will focus on male descendants of Jefferson’s male ancestors. Below is the tree starting with his great grandfather, Thomas Jefferson Senior (our Thomas Jefferson is the one in red).
1. Thomas Jefferson Sr (1653-1697) & Mary Branch (1660)
2. Thomas Jefferson Jr,
(1677-1730) & Mary Field (1679-1715)
3. Judith Jefferson
(1698-1723)
3. Thomas Jefferson III
(1700-1723)
3. Field Jefferson
(1702-1765)
3. Alice Jefferson (1704)
3. Peter Jefferson (1708-1757) &
Jane Randolph (1719-1776)
4. Jane Jefferson
(1740-1765)
4. Mary Jefferson
(1741-1804)
4. Thomas Jefferson (1743-1826) & Martha
Wayles Skelton (1748-1782)
5. Martha
"Patsy" Jefferson (1772-1836) & Thomas M Randolph
5. Jane Jefferson
(1774-1775)
5. son
(1777-1777)
5. Mary
"Polly" Jefferson (1778-1804) & John Wayles Eppes (-1823)
5. Lucy Elizabeth
(1780-1781)
5. Lucy Elizabeth
(1782-1785)
4. Elizabeth
Jefferson (1744-1777)
4. Martha Jefferson
(1746-1811) & Dabney Carr (1743-1773)
5. Jane Barbara
Carr (1766-1840)
5. Lucy Carr
(1768-1803)
5. Mary
"Polly" Carr (1768)
5. Peter Carr
(1770-1815)
5. Samuel Jefferson Carr (1771-1855)
5. Ellen Carr
(1775)
5. Martha Carr
(1775)
5. Jane Carr
(1777)
4. Peter F Jefferson
(1748-1748)
4. son (1750-1750)
4. Lucy Jefferson
(1758-1810)
4. Anna S Jefferson
(1755-1828)
4. Randolph Jefferson
(1755-1815) & Anne Jefferson Lewis
3. Mary Jefferson
(1708-1755)
3. Martha Jefferson
(1712)
2. Martha Jefferson (1679)
2. Mary Jefferson (1679)
Note that Thomas’s father, Peter Jefferson, was also short on sons, but he did have two who survived beyond their first birthday – Thomas and his brother Randolph. Peter had a brother Field, and some of his living direct male descendants have been located. Testing them will give us the Jefferson family Y chromosome.
Nephews Peter and Samuel Carr (who were implicated as possibly being the father of Sally’s children) do not share a common male ancestor with Thomas, so they would have a different Y chromosome. However there are no known direct male descents of Peter or Samuel alive today, nor are there any known living direct male descendants of their father, Dabney Carr. But there are living direct male descendants of Dabney’s father, John Carr (1706-1778).
Now let’s look at Sally Hemings family tree as shown below. Note that Thomas Jefferson himself is actually in Sally’s tree because Sally and Jefferson’s wife were half sisters (an interesting fact but totally irrelevant to our DNA testing). Both Sally and Thomas are shown in red in the tree for easy recognition.
John Wayles (1715-1773) & Martha “Patsy” Eppes (1712-1748)
Martha Wayles (1748-1782)
& Thomas Jefferson (1743-1826)
John Wayles & slave Elizabeth "Betty" Hemings (1735-1807)
5 children
Sally
Hemings (1773-1836) -- slave of Thomas Jefferson
Thomas C Woodson
(1790-1879)
(connection to
Sally is based on oral tradition)
Harriet Hemings
(1795-1797)
William Beverley
Hemings (1798-1873)
Thenia Hemings (1799
- died in infancy)
Harriet Hemings
(1801-1863)
James Madison Hemings
(1805-1877)
Thomas Eston Hemings (1808-1856)
Sally had six documented children. A seventh person, Thomas Woodson, has not been documented as being an offspring of Sally’s. But oral tradition among Woodson’s descendants states that he was parented by Thomas and Sally.
By the 1990s DNA testing had matured to the point that it could be used to unravel the 200-year old mystery. Here is a chronology of what happened next:
1998: DNA test
DNA tests were done on direct male descendants of the following people:
Field Jefferson (TJ’s uncle)
John Carr (grandfather of Peter and Samuel)
Thomas Woodson (alleged first child of Sally)
Eston Hemings (documented youngest child of Sally)
The result was that Jefferson’s Y chromosome, as obtained from descendants of Field, were found in descendants of Eston. Furthermore, Carr’s Y chromosome was not found in descendants of Eston. And, Jefferson’s Y chromosome was not found in the descendants of Woodson.
This last fact proves that Woodson was not fathered by Jefferson. He might still have been a child of Sally’s from a different father, but since the oral tradition says that he was the child of Sally and Thomas, the allegation that he is even Sally’s child is now in question.
The researchers who performed this DNA study concluded that most-probably Thomas Jefferson fathered Eston.
2000: Thomas Jefferson Memorial Foundation study
A study was done by the Thomas Jefferson Memorial Foundation (a.k.a. the Thomas Jefferson Foundation). This is the organization that runs Monticello. They looked at a broad body of evidence beyond the DNA. For example, Jefferson who traveled frequently, was always in residence when each of Sally’s six children were conceived. Also there was a strong resemblance between some of Sally’s descendants and the Jeffersons. And all of Sally’s children were conceived after Jefferson’s wife died. This study concluded that it is very unlikely that any Jefferson other than Thomas fathered Hemings six documented children.
2001: Thomas Jefferson Heritage Society study
A study was done by the hastily-formed Thomas Jefferson Heritage Society. It used the same data as was used by the Thomas Jefferson Memorial Foundation in the 2000 study. But it reached a different conclusion. It stated that the relation between Thomas Jefferson and Sally Hemings was by no means proven. And furthermore, it suggested that the most likely alternate was Thomas’s brother Randolph (although Randolph’s name was never mentioned throughout history as being a candidate).
2001: National Genealogical Society article
A second study was done in 2001, and this time by genealogists. The findings were published in the National Genealogical Society’s quarterly journal. The authors said that the link between Jefferson and Hemings was credible. They said it was consistent with the weight of the evidence. Furthermore, they went on to criticize the Thomas Jefferson Heritage Society’s report for (1) weakness in approach, (2) bias toward data, and (3) ignoring the weight of evidence.
OK, now you have it – disagreement among the studies. So I’ll let you draw your own conclusion.
c. The Anastasia Mystery
The Romanov family ruled Russia for over 300 years. They were overthrown in the Russian revolution of 1917, and the royal family was placed under house arrest. In 1918 Lenin ordered the Red Guard to kill the royal family. Those murdered were:
Tsar Nicholas II
his wife Tsarina Alexandra
his four daughters – Olga, Tatiana, Maria, and Anastasia
his son Alexei
a physician who had the misfortune of being present
three female servants
The location of the burial site was never disclosed.
In 1920 a woman was found in Berlin, claimed amnesia and took the name of Anna Anderson. In 1922 Anna Anderson revealed that she was the youngest daughter, Anastasia, and that she escaped the massacre. Her critics were skeptical and claimed that she was Franziska Schanzkowska, a Polish factory worker. When Anna Anderson died in 1984, her true identity was still unproven.
In 1991 a burial site was discovered in the Ural mountains, not far from where the massacre allegedly took place. Nine bodies were exhumed from the site. They consisted of two adult males, four adult females, and three younger females. Testing of nuclear DNA (i.e., the chromosomes) revealed that one of the adult males was closely related to the three younger females, and that one of the adult females was closely related to the three younger females. The conclusion was that they were a family group consisting of a father, mother, and three daughters. So could the nine bodies be the Tsar, Tsarina, three daughters, the physician, and the three female servants? And, if so, what happened to the fourth daughter and to the son?
It’s time to look at family trees. We’ll start with a tree of the British royal family – the House of Windsor. Here is the tree from Queen Victoria down to the present-day Queen Elizabeth.
1. Queen Victoria of England & Prince Albert
2. King Edward VII of England & Alexandra
3.King George V of England & Mary
4. King George VI of England
5. Queen Elizabeth II of England & Prince Philip Mountbatten
But why are we looking at the British royal family when we are trying to solve a mystery with the Russian royal family? That’s because Queen Victoria had another child, Alice, who was the mother of the Tsarina Alexandra. This is shown below.
1. Queen Victoria of England & Prince Albert
2. Princess Alice Maud Mary & Grand Duke Louis IV of Hesse
3. Tsarina Alexandra & Tsar Nicholas II Romanov
4. Grand Duchess Olga
4. Grand Duchess Tatiana
4. Grand Duchess Maria
4. Grand Duchess Anastasia
4. Tsarevich Alexei
2. King Edward VII of England & Alexandra
3.King George V of England & Mary
4. King George VI of England
5. Queen Elizabeth II of England & Prince Philip Mountbatten
To solve the mystery, DNA was tested from Prince Philip, husband of Queen Elizabeth. But why Philip – he married into the royal family so why would we expect him to have any of the royal DNA? That’s because Alice had another daughter, Victoria, who was the grandmother of Philip. So Philip and Elizabeth are both descended from Victoria in their own way, and were related (third cousins) before they became husband and wife. This is shown in the tree below.
1. Queen Victoria of England & Prince Albert
2. Princess Alice Maud Mary & Grand Duke Louis IV of Hesse
3. Princess Victoria of Hesse & Prince Louis of Battenberg
4. Princess Alice of Battenberg & Prince Andrew of Greece
5. Prince Philip Mountbatten & Queen Elizabeth II of England
3. Tsarina Alexandra & Tsar Nicholas II Romanov
4. Grand Duchess Olga
4. Grand Duchess Tatiana
4. Grand Duchess Maria
4. Grand Duchess Anastasia
4. Tsarevich Alexis
2. King Edward VII of England & Alexandra
3.King George V of England & Mary
4. King George VI of England
5. Queen Elizabeth II of England & Prince Philip Mountbatten
Now it’s time to follow the mitochondria. Recall that it is passed from mother to all her children, but only her female children pass it to their children. The tree below shows all people who have Victoria’s mtDNA.
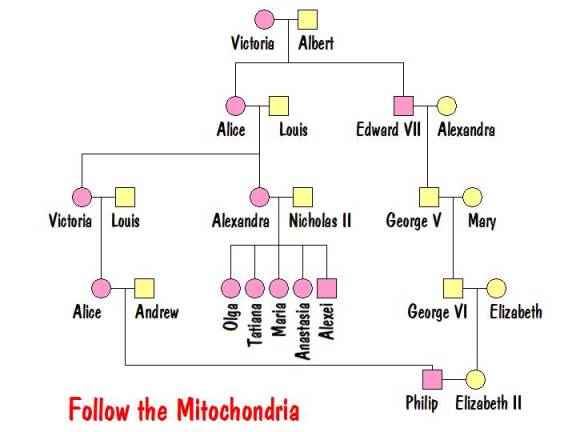
From this it is clear that Philip would have the same mtDNA as the Tsarina and all her children. And he would be the only descendant of Victoria alive today who has that mtDNA. So Philip was tested to determine if his mitochondria matched that of the exhumed family. Recall, however, that mtDNA mutates very slowly and for that reason is usually not used to determine recent ancestors. But Prince Philip’s mtDNA exhibited a rare mutation (at markers 16111 and 16537). The exhumed family had the same rare mutation. This proved that the exhumed bodies are the Romanovs.
An mtDNA match on the Tsar’s side makes the case stronger yet. The mtDNA of the exhumed body of the alleged Tsar contained an even rarer anomaly – namely two DNA values at one marker (called a heteroplasmy). Since the location of the Tsar’s brother’s grave was known, he was exhumed and tested. It contained the same heteroplasmy.
There was now no question that the remains were that of the royal family. And the missing son and daughter gave credence to Anna Anderson’s claim of being Anastasia.
The only way to solve the Anastasia mystery now was to test Anna Anderson’s DNA. But she had died seven years before the gravesite was discovered. Fortunately her DNA had been preserved. One report I read said that it came from a biopsy sample wrapped in paraffin and stored in a hospital freezer. Another said it was from a lock of hair that her husband saved. From whichever source, we now had a sample of her mtDNA for comparison purposes. And the results came back very conclusively. IT DID NOT MATCH!!! ANDERSON WAS A FRAUD. However Anderson’s mtDNA did match that of Karl Maucher, maternal great nephew of Polish factory worker Franziska Schanzkowska.
But what did happen to Anastasia and Alexei? Would another Anna Anderson come forward in the future to claim the title. Well in 2007 another burial site was discovered, close to the first one. It contained the bodies of a young boy and a young girl. DNA tests revealed that they were from the same family as the bodies at the other site, so they must be Anastasia and Alexei. MYSTERY SOLVED!!!
7. GENETIC DISEASES (DOWN, SICKLE CELL, TAY-SACHS,
HEMOPHILIA)
Although there are many genetic diseases, the above four have been singled out for discussion here because they are each well known, and they each illustrate a different genetic point.
a. Down Syndrome
This disease is found in all populations and doesn’t favor any particular one. It is a chromosomal disorder rather than a genetic one, and involves an extra copy of chromosome 21. Two copies of the chromosome are obtained from one parent (usually the mother) and one from the other. Since it is usually the mother who gives the extra chromosome, it is not surprising that a characteristic of the mother (her age) might be a factor.
b. Sickle Cell Anemia
This disease is most prevalent in Sub-Saharan African populations. It is due to a mutation in the beta-globin gene on chromosome 11. The particular mutation is a SNiP whereby a GAG sequence becomes a GTG, resulting in amino acid glutamate instead of valine. The mutant gene is recessive, meaning that a person needs to have two mutant genes in order to have the disease.
Since the disease is so deadly, it would seem that it would eventually become eradicated simply by survival of the fittest. That would be the case except for another factor involved here. Namely having a single mutant gene gives protection against malaria. So in regions where malaria is prevalent, those who have two good genes have a better chance of dying from malaria, leaving those with at least one mutant gene alive and thriving. This is referred to as a heterozygote advantage – a fancy term meaning that it’s better to be a carrier of the disease than not.
c. Tay-Sachs Disease
This disease is prevalent in three distinct populations – Ashkenazi Jews, Louisiana Cajuns, and French Canadians. The disease is linked to the HEXA gene on chromosome 15 and is recessive. Any mutation that causes the HEXA gene to produce a different protein will result in the disease.
There are over 90 different identified mutations of the HEXA gene. Some are SNiPs, some are STiRs, and there are other types of mutations as well. The particular mutation that causes the disease in the Ashkenazi Jewish population is a STiR resulting in an extra repeat of TATC. Since every three base pairs define a particular amino acid, inserting four extra base pairs changes not just the current amino acid but alters all following amino acids in the sequence as well. The French Canadian mutation is a so-called long sequence deletion. So there is no relation between the Ashkenazi Jews having the disease and the French Canadians having it – the original mutations occurred in two different people. What about the Louisiana Cajuns? You would think that they would have inherited it from the same original person as the French Canadians – after all, both populations have a French connection. But, surprisingly, the Louisiana Cajun mutation is the same as the Ashkenazi Jewish one. Makes you wonder if perhaps there was a Jewish fur trader in New Orleans.
d. Hemophilia
Hemophilia is more prevalent among men than among women. It is caused by a mutant F8 or F9 gene (there are two types of hemophilia) on the X chromosome. And it is recessive.
Since women have two X chromosomes, they would need to have the mutation in both of their Xs in order to have the disease. Men, on the other hand, have only one X chromosome. So if that one chromosome has the mutation, they don’t have the possibility of a good X chromosome to dominate over the mutant one. This explains why the disease is more prevalent in men.
The same applies to colorblindness. It too is linked to the X chromosome, and is more prevalent in men than in women.
8. APPLICATIONS OF DNA (IDENTITY, PATERNITY, GENEALOGY)
This section summarizes the uses of DNA that we have seen so far, and introduces a few new ones.
a. Human Migration Patterns and Population Studies
Involves slowly-changing markers in the junk region of the Y chromosome or mtDNA.
b. Genealogy
Involves rapidly-changing markers in junk region of Y chromosome.
c. Genetic Testing for Characteristics (Diseases)
Involves markers in actual genes (not junk) on specific chromosomes.
d. Paternity
Testing
Involves testing values on the child and subtracting out those that are present in the mother. The remaining values had to come from the father, so any male not possessing those values can be ruled out.
e. Forensic Identity Checking (FBI CODIS database)
Involves testing of rapidly-changing markers in junk regions across various chromosomes. By having enough markers, they are able to build up a “fingerprint” unique to only one individual.
9. EPILOG
That just about covers all I’ve learned about this topic. I’ve found it fascinating to realize how DNA can be used to tell us something about our very early ancestors, and also to determine if we share a recent common ancestor with some other person. It’s all locked up in those As, Cs, Gs, and Ts.
I hope I’ve been successful in passing this knowledge on to you. Armed with this information, you should now be able to hold your head up high when you attend some of the DNA lectures given at genealogy conferences.